Discovery of the Phage Nucleus and PhuZ Spindle.
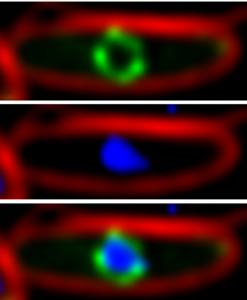 |
| The phage nucleus of jumbo Pseudomonas phage forms during lytic growth. A shell of protein (green) formed by gp105 surrounds replicating phage 201phi2-1 DNA (blue) inside a Pseudomonas chlororaphis cell (red cell membranes). Top: GFP-gp105 (green), Middle: DNA (blue), Bottom, merged. |
Our lab has been working on the cell biology of Pseudomonas ΦKZ-like phage for more than a decade. We identified and characterized a family of divergent tubulin-related proteins encoded by “jumbo” bacteriophage that infect Pseudomonas species that we termed “PhuZ” (Phage TubZ-like) in collaboration with David Agard at UCSF. This led to the discovery of a new paradigm for phage reproduction (Kraemer et al. Cell 2012; Erb Curr Opin Microbiol 2013). As we investigated further, we observed that after injection of the phage genome into the host cell, the DNA migrates to the center of the cell, forming a centrally localized compartment. Migration of phage DNA to midcell depends upon PhuZ, which forms triple-stranded filaments that assemble a bipolar spindle (Kraemer et al. Cell 2012; Zher et al. Structure 2014). PhuZ filaments display dynamic instability in vivo and in vitro (Erb et al. eLife 2014). This represents the first identification of a prokaryotic tubulin with the properties of microtubules and the ability to form a bipolar spindle.
This led to the discovery that certain phage form a nucleus-like structure during lytic growth we termed the “phage nucleus” (Chaikeeratisak et al. Science 2017; phagenucleus.org). We initially focused on Pseudomonas phage, including ΦPA3, ΦKZ, and 201φ2-1, and characterized their phage nuclei, which physically separate viral DNA from the cytoplasm (Chaikeeratisak et al. Cell Reports 2017, Laughlin & Deep et al. Science 2022). The phage nucleus assembles from protomers of a phage-encoded nuclear shell protein we termed “chimallin” (after the ancient Aztec shield chimalli, as the phage nucleus shields the phage genome from host defenses) immediately after DNA injection, grows larger as the DNA replicates, and is positioned at midcell by the bipolar PhuZ spindle. Protein localization data show that DNA replication and transcription occur inside the phage nucleus, while translation and metabolic processes, such as nucleotide synthesis, occur outside of the nucleus. Time-lapse microscopy demonstrates that proteins destined for the nucleus are synthesized in the cytoplasm and then accumulate in the phage nucleus. This suggests a selective two-way exchange of mRNA, protein, and nucleotides between the cytoplasm and nucleus, as in eukaryotes. Late in infection, viral capsids assemble on the cell membrane and traffic through the cell along treadmilling PhuZ filaments (Chaikeeratisak et al. Cell 2019). Capsids dock on the surface of the phage nucleus, where DNA packaging occurs. Ultimately, fully assembled viral particles form, sometimes into ordered structures called “bouquets” (Chaikeeratisak et al. Sci Adv 2022), and are released when the cell lyses. These results demonstrate that bacteriophage evolved both a spindle and a nucleus, and provide insight into the evolution of these defining structures of eukaryotic cells.
We have come a long way since the identification of four Pseudomonas phages that encode PhuZ: we now know of over 100 phage (including non-jumbo phages as small as 167kb) encoding chimallin homologs that are all predicted to replicate using this same pathway. We have found that the phage nucleus and spindle can act as phage speciation factors (Chaikeeratisak et al. Nat Commun 2021). We have started characterizing divergent nucleus-forming phage that infect hosts outside of the pseudomonads (Birkholz et al. Cell Rep 2022). And we have started to probe questions on how import and export through the phage nuclear shell works and how we can begin to use genetic tools to manipulate nucleus forming phages and further study the exciting cell biology of their infection cycles (Nguyen et al. PLoS One 2021).
Antibiotic discovery and mechanism of action.
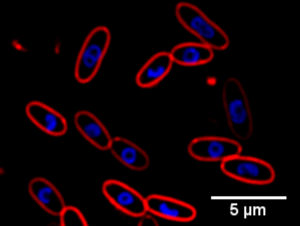 |
| BCP allows the rapid identification of antibiotic mechanism of action. In the example above, the DNA (blue) of E. coli cells (red cell membranes) forms toroidal structures when treated with protein synthesis inhibitors. |
Antibiotic resistance among bacteria is a growing public health concern. Once easily treated with penicillin, infections caused by methicillin-resistant Staphylococcus aureus (MRSA) and vancomycin-resistant Enterococcus (VRE) are now very difficult to combat. Cases of highly drug resistant Gram-negative bacteria are becoming increasingly common throughout the world. Without effective antibiotics for these drug resistant pathogens, even countries with well-developed health care systems will have few treatment options. Therefore, the need for new treatments that are active against multidrug resistant pathogens is more urgent than ever before. Unfortunately, the number of new antibiotics being brought to market is currently insufficient to keep pace with the rate of antibiotic resistance development.
To combat the imminent antibiotic resistance crisis, our lab has an ongoing focus on finding cures for multidrug resistant infections. We study some of the latest antibiotics to enter the clinic as well as preclinical molecules and natural products to determine their mechanisms of action (Nonejuie et al. J Antibiot 2016). We investigate how resistance to these new antibiotics arises and how treatment can be modified to lead to a more successful clinical outcome (Poudel et al. Sci Data 2019; Rajput et al. Sci Data 2019; Rajput et al. Gigascience 2021). We have developed bacterial cytological profiling (BCP) in collaboration with Kit Pogliano, a new method for screening for novel antibiotics and determining their mechanisms of action (Nonejuie et al. PNAS 2013; Lamsa et al. Mol Micro 2012). We are using this method to screen libraries of natural products for novel compounds active against multidrug resistant bacteria. BCP can be used to screen for novel chemical scaffolds with activity against new targets in bacteria, to determine antibiotic mechanism of action, and for rapid antibiotic susceptibility testing (Nonejuie et al. PNAS 2013; Lamsa et al. ACS Chem Biol 2016; Peters et al. ACS Chem Biol 2018; Montaño et al. J Bacteriol 2021). To accomplish these goals, we have set up collaborations with clinicians at the UCSD School of Medicine, with pharmaceutical companies who have promising new antibiotics, and with chemists who can provide novel compounds for screening for new antibiotics. We are also working to understand how combinations of antibiotics can be used in the case of antibiotic resistant bacteria to lead to a more successful clinical outcome (Sakoulas et al. AAC 2013; Werth et al. AAC 2013; Sakoulas et al. AAC 2012; Barber et al. AAC 2014; Sakoulas et al. AAC 2015). In addition, undergraduate students enrolled in the Phage Hunters Advancing Genomics and Evolutionary Science (PHAGE) class isolated Streptomyces strains with novel biosynthetic clusters and inhibitory activity against MRSA (Montaño et al. PLoS One 2022).
Currently, we are collaborating with Elizabeth Villa, Kevin Corbett, David Pride, Justin Meyer, Kit Pogliano, Paul Turner (Yale), and John Glass (JCVI) as part of the HHMI Emerging Pathogens Initiative in an effort to find treatments for multidrug resistant bacterial infections. This collaboration will bring antibiotics and phage research together by investigating phage therapy opportunities and how phage and antibiotics can work synergistically to combat multidrug resistant infections.